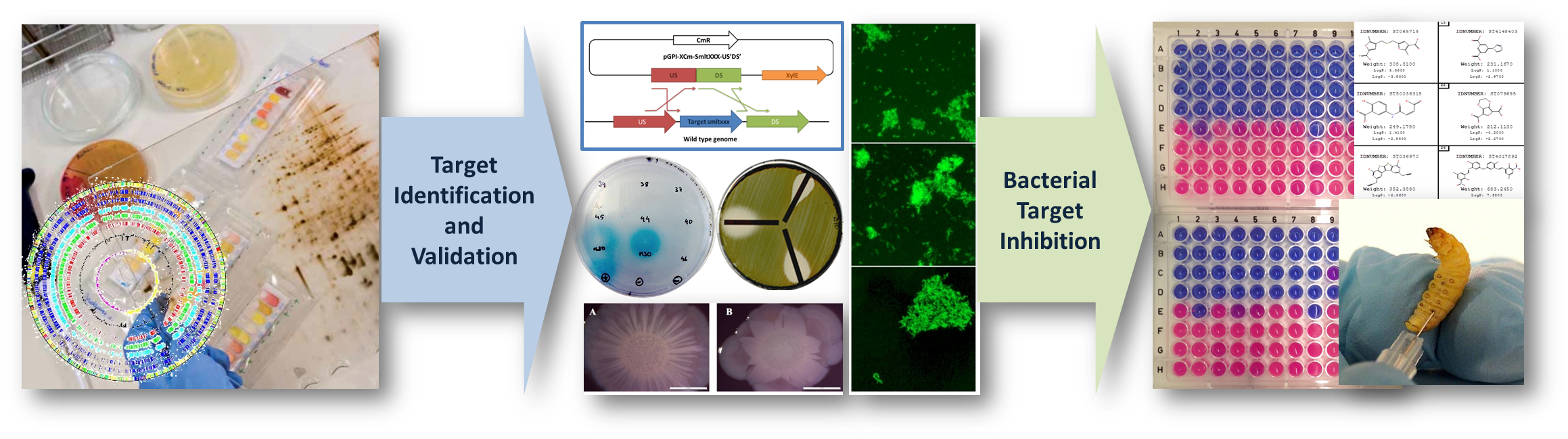
QUORUM-SENSING-DEPENDENT RESPONSE REGULATORS IN STENOTROPHOMONAS MALTOPHILIA AS SOURCE OF NOVEL ANTIMICROBIAL AND ANTIBIOFILM STRATEGIES (STENORR). Convocatòria PID 2023; Ministerio de Ciencia e Innovación (MICINN). Execució: 1/09/24 → 31/08/27
QUORUM SENSING REGULATORY NETWORKS IN STENOTROPHOMONAS MALTOPHILIA AS TARGETS FOR THERAPEUTIC ALTERNATIVES TO CURRENT ANTIBIOTICS. Convocatòria PID2019; Ministeri de Ciència i Innovació (MICINN). Execució: 1/06/20 → 30/05/23
The Mla system and its role in maintaining outer membrane barrier function in Stenotrophomonas maltophilia. Front Cell Infect Microbiol. 2024 Feb 26;14:1346565. doi: 10.3389/fcimb.2024.1346565.
Synthesis and evaluation of aromatic BDSF bioisosteres on biofilm formation and colistin sensitivity in pathogenic bacteria. Eur J Med Chem. 2023 Dec 5;261:115819. doi: 10.1016/j.ejmech.2023.115819.
A Stenotrophomonas maltophilia TetR-Like Transcriptional Regulator Involved in Fatty Acid Metabolism Is Controlled by Quorum Sensing Signals. Appl Environ Microbiol. 2023 Jun 28;89(6):e0063523. doi: 10.1128/aem.00635-23.
Improved mini-Tn7 Delivery Plasmids for Fluorescent Labeling of Stenotrophomonas maltophilia. Appl Environ Microbiol. 2023 Jun 28;89(6):e0031723. doi: 10.1128/aem.00317-23.
Synthesis and evaluation of novel furanones as biofilm inhibitors in opportunistic human pathogens. Eur J Med Chem. 2022; 242:114678. doi: 10.1016/j.ejmech.2022.114678.
Strain-specific interspecies interactions between co-isolated pairs of Staphylococcus aureus and Pseudomonas aeruginosa from patients with tracheobronchitis or bronchial colonization. Sci Rep. 2022; 12(1):3374. doi: 10.1038/s41598-022-07018-5.
The Pseudomonas aeruginosa substrate-binding protein Ttg2D functions as a general glycerophospholipid transporter across the periplasm. Commun Biol. 2021; 4(1):448. doi: 10.1038/s42003-021-01968-8.
Antigen Discovery in Bacterial Panproteomes. Methods Mol Biol. 2021; 2183:43-62. doi: 10.1007/978-1-0716-0795-4_5.
Effects of cigarette smoke on the administration of isoniazid and rifampicin to macrophages infected with Mycobacterium tuberculosis. Exp Lung Res. 2021; 47(2):87-97. doi: 10.1080/01902148.2020.1854371.
Phenotypic and Transcriptomic Analyses of Seven Clinical Stenotrophomonas maltophilia Isolates Identify a Small Set of Shared and Commonly Regulated Genes Involved in the Biofilm Lifestyle. Appl Environ Microbiol. 2020; 86(24):e02038-20. doi: 10.1128/AEM.02038-20.
Genetic Variants of the DSF Quorum Sensing System in Stenotrophomonas maltophilia Influence Virulence and Resistance Phenotypes Among Genotypically Diverse Clinical Isolates. Front Microbiol. 2020; 11:1160. doi: 10.3389/fmicb.2020.01160.
The phylogenetic landscape and nosocomial spread of the multidrug-resistant opportunist Stenotrophomonas maltophilia. Nat Commun. 2020; 11(1):2044. doi: 10.1038/s41467-020-15123-0.
E-cigarettes: Effects in phagocytosis and cytokines response against Mycobacterium tuberculosis. PLoS One. 2020; 15(2):e0228919. doi: 10.1371/journal.pone.0228919.
Sulfonamide-based diffusible signal factor analogs interfere with quorum sensing in Stenotrophomonas maltophilia and Burkholderia cepacia. Future Med Chem. 2019 Jul;11(13):1565-1582. doi: 10.4155/fmc-2019-0015.
Heterogeneous Colistin-Resistance Phenotypes Coexisting in Stenotrophomonas maltophilia Isolates Influence Colistin Susceptibility Testing. Front Microbiol. 2018; 9:2871. doi: 10.3389/fmicb.2018.02871.
Quorum Sensing Signaling and Quenching in the Multidrug-Resistant Pathogen Stenotrophomonas maltophilia. Front Cell Infect Microbiol. 2018; 8:122. doi: 10.3389/fcimb.2018.00122.
Proteomic analysis of outer membrane proteins and vesicles of a clinical isolate and a collection strain of Stenotrophomonas maltophilia. J Proteomics. 2016; 142:122-9. doi: 10.1016/j.jprot.2016.05.001.
Triclosan-induced genes Rv1686c-Rv1687c and Rv3161c are not involved in triclosan resistance in Mycobacterium tuberculosis. Sci Rep. 2016; 6:26221. doi: 10.1038/srep26221.
Decoding the genetic and functional diversity of the DSF quorum-sensing system in Stenotrophomonas maltophilia. Front Microbiol. 2015; 6:761. doi: 10.3389/fmicb.2015.00761. eCollection 2015.
Draft Genome Sequence of Stenotrophomonas maltophilia Strain UV74 Reveals Extensive Variability within Its Genomic Group. Genome Announc. 2015; 3(3):e00611-15. doi: 10.1128/genomeA.00611-15.
Stenotrophomonas maltophilia responds to exogenous AHL signals through the LuxR solo SmoR (Smlt1839). Front Cell Infect Microbiol. 2015; 5:41. doi: 10.3389/fcimb.2015.00041.
Animals devoid of pulmonary system as infection models in the study of lung bacterial pathogens. Front Microbiol. 2015; 6:38. doi: 10.3389/fmicb.2015.00038.
Draft Genome Sequence of Stenotrophomonas maltophilia Strain M30, Isolated from a Chronic Pressure Ulcer in an Elderly Patient. Genome Announc. 2014; 2(3):e00576-14. doi: 10.1128/genomeA.00576-14.
Two different rpf clusters distributed among a population of Stenotrophomonas maltophilia clinical strains display differential diffusible signal factor production and virulence regulation. J Bacteriol. 2014; 196(13):2431-42. doi: 10.1128/JB.01540-14.
antibacTR: dynamic antibacterial-drug-target ranking integrating comparative genomics, structural analysis and experimental annotation. BMC Genomics. 2014; 15(1):36. doi: 10.1186/1471-2164-15-36.
Abundance of the Quorum-Sensing Factor Ax21 in Four Strains of Stenotrophomonas maltophilia Correlates with Mortality Rate in a New Zebrafish Model of Infection. PLoS One. 2013; 8(6):e67207. doi: 10.1371/journal.pone.0067207.